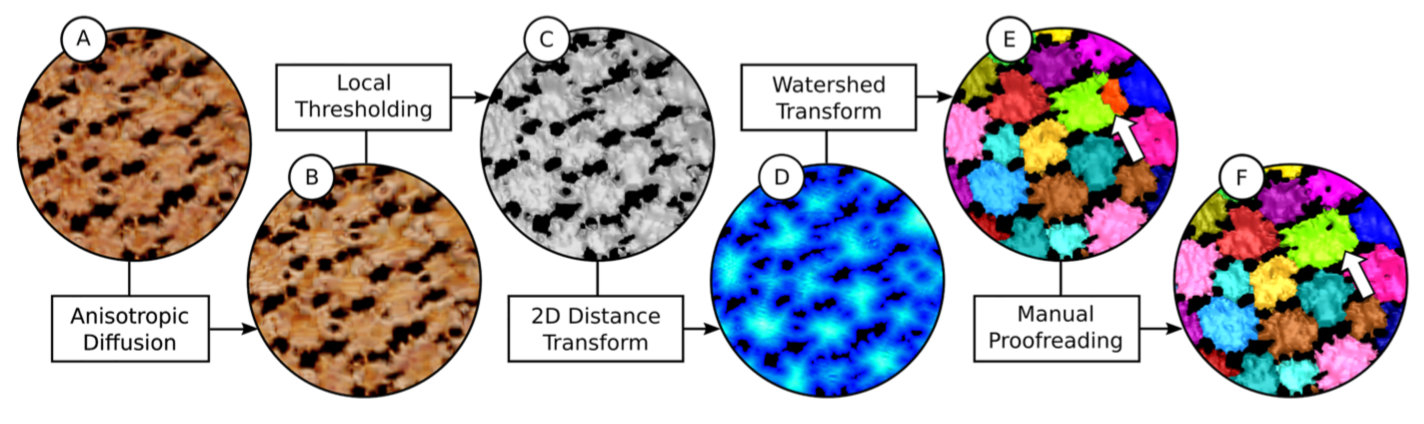
Automated segmentation of complex patterns in biological tissues
Many biological structures show recurring tiling patterns on one structural level or the other. Current image acquisition techniques (e.g. Micro computed tomography) are able to resolve those tiling patterns to allow quantitative analyses. The resulting image data, however, may contain an enormous number of elements. This renders manual image analysis infeasible, in particular when statistical analysis is to be conducted, requiring a larger number of image data to be analyzed. As a consequence, the analysis process needs to be automated to a large degree. In Knötel et al. 2017 we describe a multi-step image segmentation pipeline for the automated segmentation from computed tomography data of an examplary natural tiling, called tessellated cartilage, occuring in the cartilaginous skeletons of sharks and rays ( Knötel et al. 2017. Automated segmentation of complex patterns in biological tissues: Lessons from stingray tessellated cartilage." PloS one 12.12 (2017): e0188018 ).
The endoskeleton of sharks and rays is comprised of a cartilaginous core, covered by thousands of mineralized tiles, called tesserae. Characterizing the relationship between tesseral morphometrics, skeletal growth and mechanics is challenging because tesserae are small (a few hundred micrometers wide), anchored to the surrounding tissue in complex three-dimensional ways, and occur in huge numbers. We use µCT scans of skeletal elements from an ontogenetic series of round stingray
Urobatis halleri and this custom-made automatic segmentation algorithm for a first quantitative analysis of the tesserae network to gain valuable insights into the generation and maintenance of elasmobranch tessellated cartilage (article coming soon).